Note
Click here to download the full example code
Extend scikit-learn FastICA example to use Icasso¶
FastICA is fit multiple times to simple example data and performance can be visually inspected.
# Authors: Erkka Heinila <erkka.heinila@jyu.fi>
#
# License BSD (3-clause)
from itertools import cycle
import numpy as np
import matplotlib.pyplot as plt
from scipy import signal
from sklearn.decomposition import FastICA
from sklearn.decomposition import PCA
from icasso import Icasso
For replicability
random_state = 50
distance=0.12
Generate sample data
n_samples = 2000
time = np.linspace(0, 8, n_samples)
s1 = np.sin(2 * time) # Signal 1 : sinusoidal signal
s2 = np.sign(np.sin(3 * time)) # Signal 2 : square signal
s3 = signal.sawtooth(2 * np.pi * time) # Signal 3: saw tooth signal
S = np.c_[s1, s2, s3]
Add noise and standardize
S += 0.4 * np.random.RandomState(random_state).normal(size=S.shape)
S /= S.std(axis=0)
Mix data
A = np.array([[1, 1, 1], [0.5, 2, 1.0], [1.5, 1.0, 2.0]])
X = np.dot(S, A.T)
Define functions for extracting bootstraps and unmixing matrices from ica object
def bootstrap_fun(data, generator):
return data[generator.choice(range(data.shape[0]), size=data.shape[0]), :]
def unmixing_fun(ica):
return ica.components_
Create the Icasso object
ica_params = {
'n_components': 3
}
icasso = Icasso(FastICA, ica_params=ica_params, iterations=100, bootstrap=True,
vary_init=True)
Fit the Icasso
icasso.fit(data=X, fit_params={}, random_state=random_state,
bootstrap_fun=bootstrap_fun, unmixing_fun=unmixing_fun)
Plot dendrogram
icasso.plot_dendrogram()
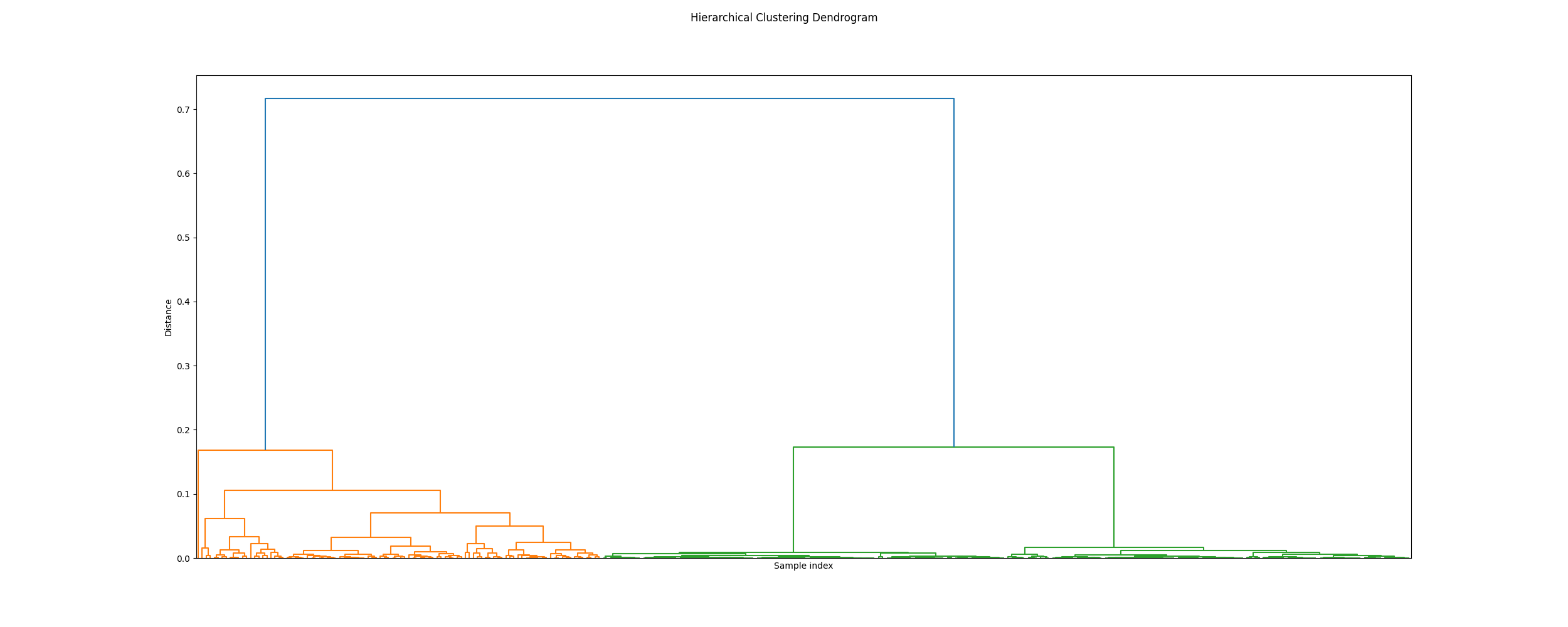
Out:
<Figure size 2500x1000 with 1 Axes>
Plot mds
icasso.plot_mds(distance=distance)
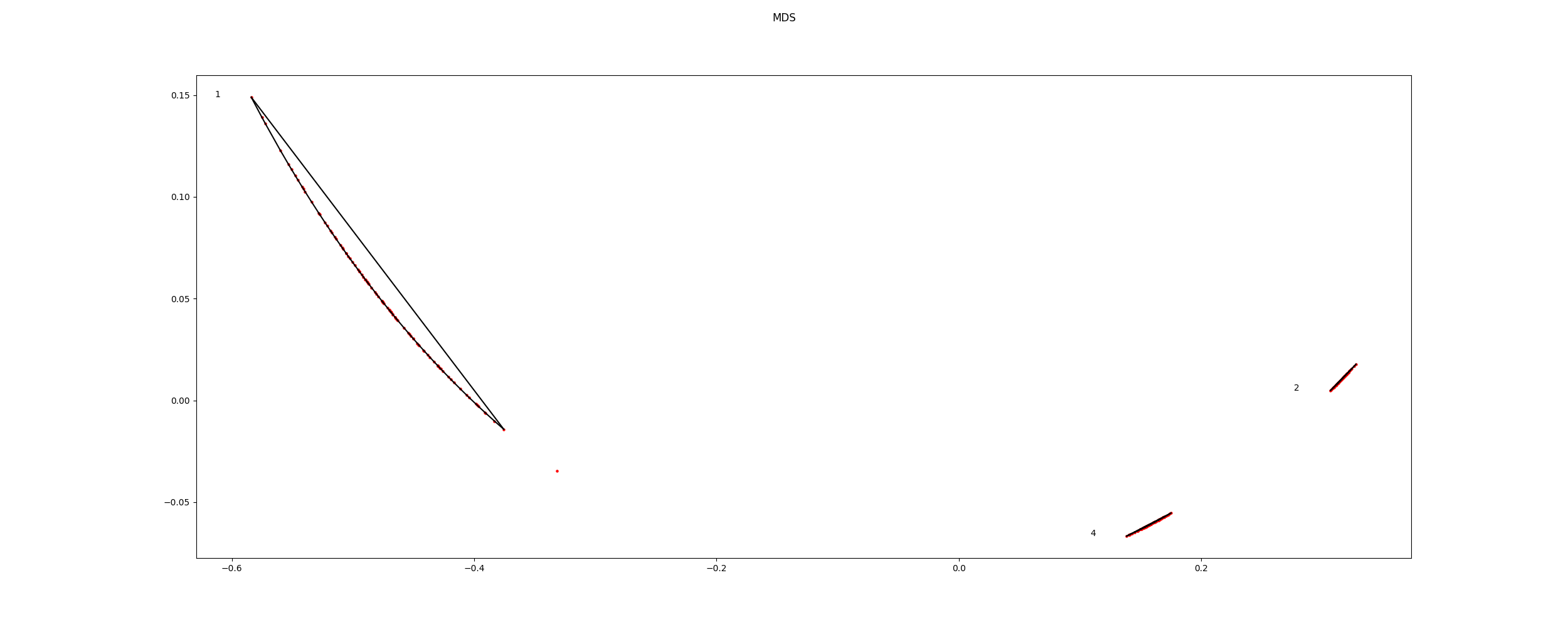
Out:
<Figure size 2500x1000 with 1 Axes>
Get the unmixing matrix and use it get the sources.
W_, scores = icasso.get_centrotype_unmixing(distance=distance)
S_ = np.dot(W_, X.T).T
# #############################################################################
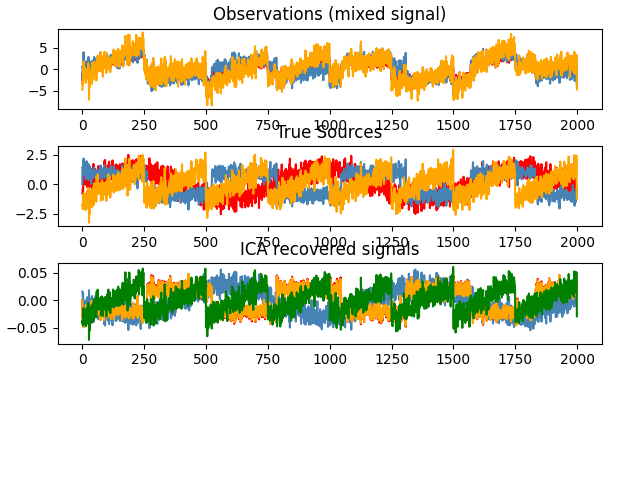
# Plot results
plt.figure()
models = [X, S, S_,]
names = ['Observations (mixed signal)',
'True Sources',
'ICA recovered signals']
for ii, (model, name) in enumerate(zip(models, names), 1):
colors = cycle(['red', 'steelblue', 'orange', 'green', 'yellow'])
plt.subplot(4, 1, ii)
plt.title(name)
for sig, color in zip(model.T, colors):
plt.plot(sig, color=color)
plt.subplots_adjust(0.09, 0.04, 0.94, 0.94, 0.26, 0.46)
plt.show()
Total running time of the script: ( 0 minutes 2.914 seconds)