Note
Click here to download the full example code
Use Icasso to compute and validate ICA on MEG data¶
ICA is fit to MEG raw data multiple times, and the performance is then visually inspected. Finally the components are retrieved as centrotypes of the most robust clusters.
# Authors: Erkka Heinila <erkka.heinila@jyu.fi>
#
# License: BSD (3-clause)
import logging
import numpy as np
import matplotlib.pyplot as plt
import mne
from mne.preprocessing import ICA
from mne.preprocessing import create_ecg_epochs, create_eog_epochs
from mne.datasets import sample
from icasso import Icasso
Set up logging
logging.basicConfig(level=logging.INFO)
Setup paths and prepare raw data.
data_path = sample.data_path()
raw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
raw = mne.io.read_raw_fif(raw_fname, preload=True)
raw.filter(1, None, fir_design='firwin')
picks = mne.pick_types(raw.info, meg='grad', eeg=False, eog=False,
stim=False, exclude='bads')
raw.drop_channels([ch_name for idx, ch_name in enumerate(raw.info['ch_names'])
if idx not in picks])
Out:
/home/erpipehe/Code/Teekuningas/icasso/examples/plot_icasso_from_raw.py:32: DeprecationWarning: data_path functions now return pathlib.Path objects which do not natively support the plus (+) operator, switch to using forward slash (/) instead. Support for plus will be removed in 1.2.
raw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
Opening raw data file /home/erpipehe/mne_data/MNE-sample-data//MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Reading 0 ... 41699 = 0.000 ... 277.709 secs...
Filtering raw data in 1 contiguous segment
Setting up high-pass filter at 1 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal highpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 1.00
- Lower transition bandwidth: 1.00 Hz (-6 dB cutoff frequency: 0.50 Hz)
- Filter length: 497 samples (3.310 sec)
Removing projector <Projection | PCA-v1, active : False, n_channels : 102>
Removing projector <Projection | PCA-v2, active : False, n_channels : 102>
Removing projector <Projection | PCA-v3, active : False, n_channels : 102>
Removing projector <Projection | Average EEG reference, active : False, n_channels : 60>
Plot raw data
raw.plot(block=True)
Out:
Using matplotlib as 2D backend.
Opening raw-browser...
<MNEBrowseFigure size 800x800 with 4 Axes>
Define parameters for mne’s ICA-object and create a Icasso object. Set bootstrap=True to use bootstrapping.
ica_params = {
'n_components':20,
'method': 'fastica',
'max_iter': 1000,
}
icasso = Icasso(ICA, ica_params=ica_params, iterations=30,
bootstrap=False, vary_init=True)
Set up params for ICA.fit method.
fit_params = {
'decim': 3,
'verbose': 'warning'
}
Set up function for getting bootstrapped versions of Raw object.
def bootstrap_fun(raw, generator):
sample_idxs = generator.choice(range(raw._data.shape[1]), size=raw._data.shape[1])
raw = raw.copy()
raw._data = raw._data[:, sample_idxs]
return raw
Set up function to get unmixing matrix from mne’s ICA object after fitting.
def unmixing_fun(ica):
unmixing_matrix = np.dot(ica.unmixing_matrix_,
ica.pca_components_[:ica.n_components_])
return unmixing_matrix
Set up function to store information about individual runs. We do this to get pca mean and pre_whiten information.
def store_fun(ica):
data = {'pre_whitener': ica.pre_whitener_,
'pca_mean': ica.pca_mean_[:, np.newaxis]}
return data
For replicability
random_state = 10
distance = 0.75
Fit icasso to raw data.
icasso.fit(data=raw, fit_params=fit_params, random_state=random_state,
bootstrap_fun=bootstrap_fun, unmixing_fun=unmixing_fun,
store_fun=store_fun)
Out:
INFO:icasso:Fitting ICA 30 times.
Plot a dendogram
icasso.plot_dendrogram()
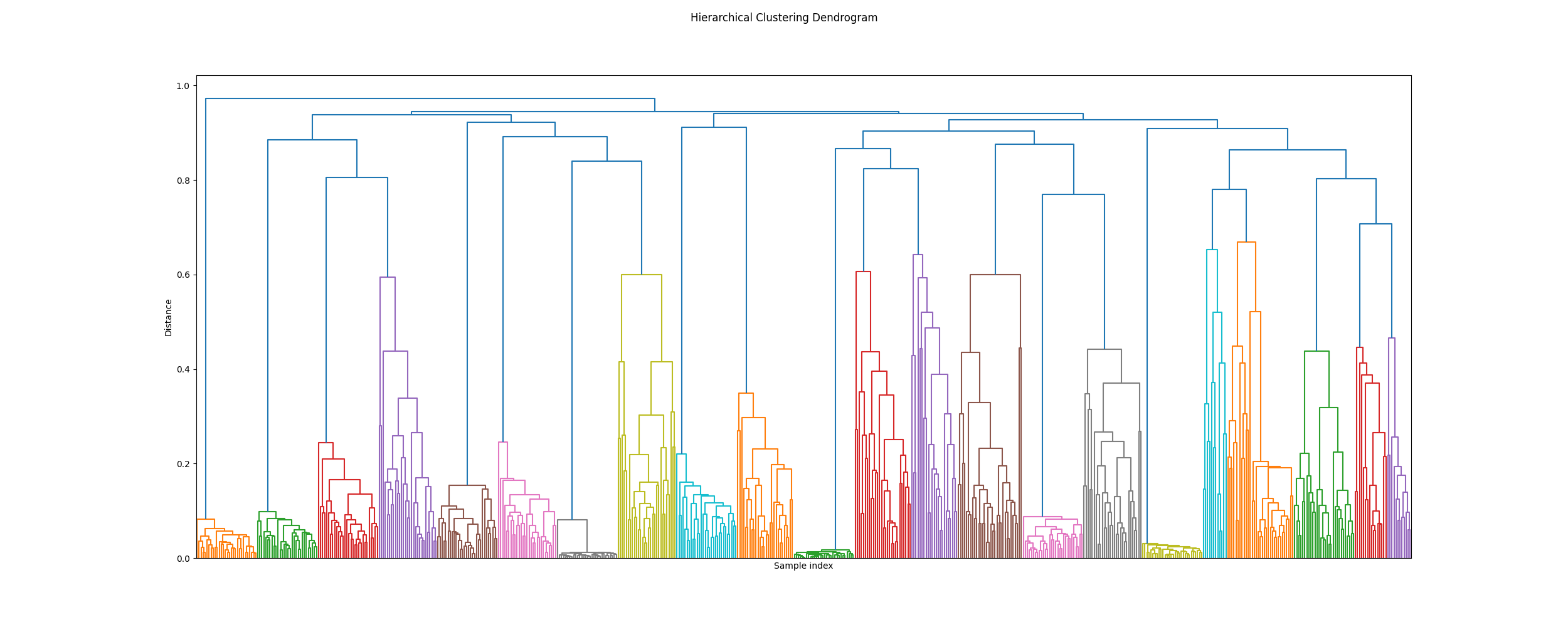
Out:
INFO:icasso:Plotting dendrogram..
<Figure size 2500x1000 with 1 Axes>
Plot the components in 2D space.
icasso.plot_mds(distance=distance)
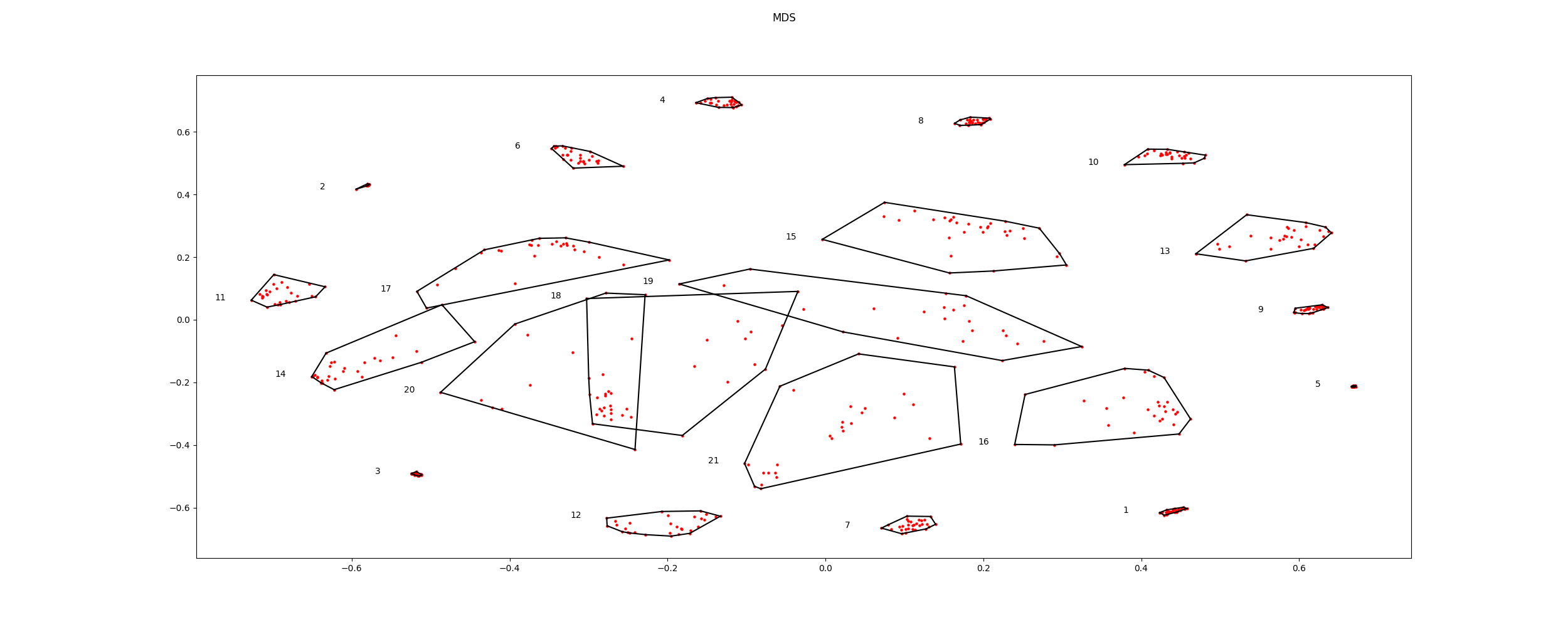
Out:
INFO:icasso:Projecting components to 2D space with MDS
INFO:icasso:Plotting ICA components in 2D space..
<Figure size 2500x1000 with 1 Axes>
Unmix using the centrotypes
unmixing, scores = icasso.get_centrotype_unmixing(distance=distance)
pca_mean, pre_whitener = icasso.store[0]['pca_mean'], icasso.store[0]['pre_whitener']
sources = np.dot(unmixing, (raw._data / pre_whitener) - pca_mean)
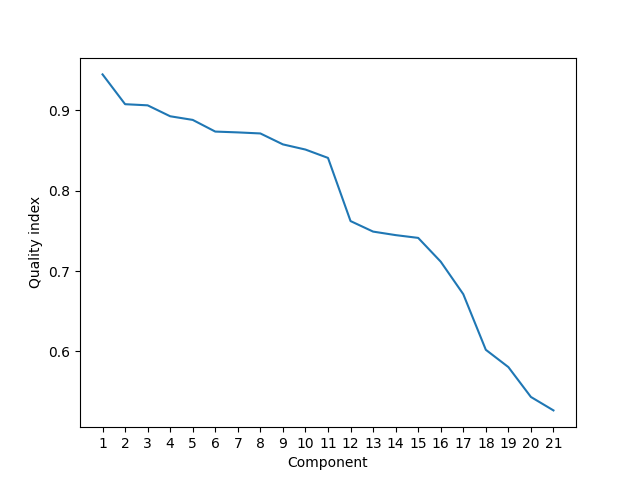
Show cluster quality indices.
plt.figure()
plt.plot(range(1, len(scores)+1), scores)
plt.xticks(range(1, len(scores)+1),
[str(idx) for idx in range(1, len(scores)+1)])
plt.xlabel('Component')
plt.ylabel('Quality index')
plt.show()
Create raw object using the icasso centrotype sources and plot it
sources = sources * 3e-04
info = mne.create_info(['ICA %03d' % (idx+1) for idx in range(sources.shape[0])],
raw.info['sfreq'], ch_types='misc')
components = mne.io.RawArray(sources, info)
components.plot(block=True)
Out:
Creating RawArray with float64 data, n_channels=21, n_times=41700
Range : 0 ... 41699 = 0.000 ... 277.709 secs
Ready.
Opening raw-browser...
<MNEBrowseFigure size 800x800 with 4 Axes>
Total running time of the script: ( 1 minutes 5.173 seconds)